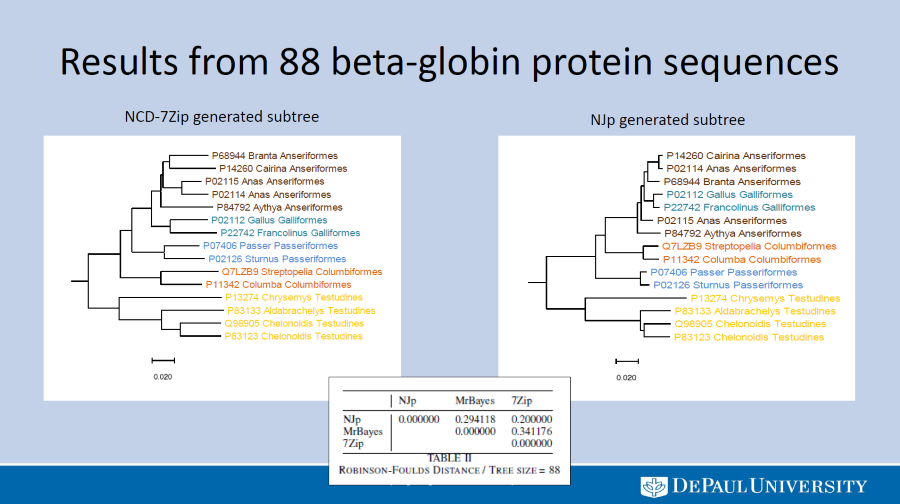
Comparing phylogeny by compression to phylogeny by NJp and Bayesian Inference
About
We compare the accuracy of the Normalized Compress Distance (NCD) technique for building phylogenetic trees from molecular data with the more widely used techniques of Neighbor joining with p-value and Bayesian inference. The NCD algorithm is a distance-based alignment-free method and as such takes unaligned sequence data as input and outputs a distance matrix. The matrix is given as input to FastME, which outputs a tree in Newick format. Every step of this process requires minimal processing and as such provides fast execution times along with minimal restrictions on input data.